How a Tiny Protein Could Revolutionize Environmental Cleanup
When you think of pollution, what comes to mind? Probably images of smog-choked cities, littered beaches, or perhaps oil spills. But there’s a class of pollutants that’s even more insidious—per- and polyfluoroalkyl substances, or PFAS. Often called “forever chemicals,” PFAS are virtually indestructible, accumulating in the environment and our bodies over time. Until recently, scientists had limited tools to combat these persistent pollutants. But now, thanks to cutting-edge research, we’re on the brink of a breakthrough that could turn the tide in the fight against these toxic chemicals.
The Hidden Power of a Simple Bacterium
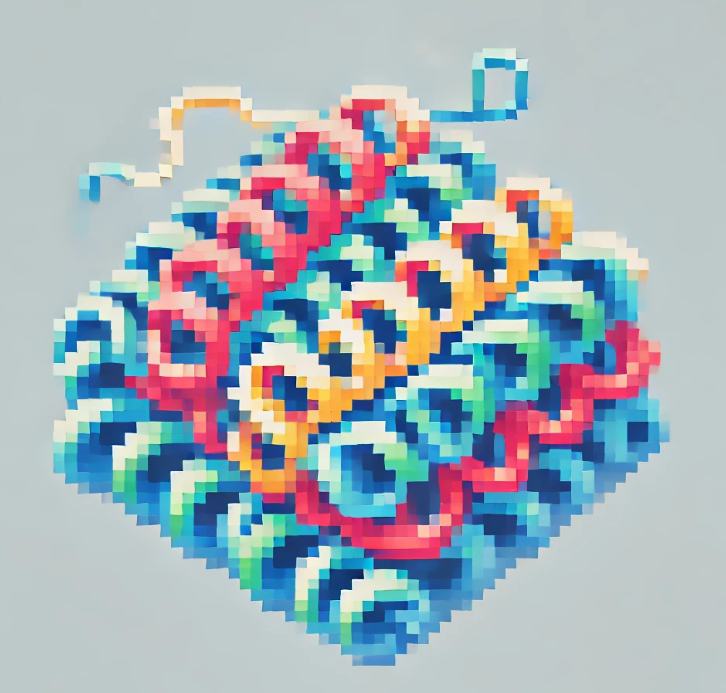
Imagine a world where tiny microorganisms help us clean up our mess. This isn’t science fiction; it’s happening right now. In a recent study, scientists explored a bacterium from the Acidimicrobiaceae family, specifically its ability to degrade PFAS. The key player? A protein known as T7RdhA.
But what makes T7RdhA so special? This protein, identified using advanced tools like AlphaFold2 (AF2), has shown a remarkable ability to break down one of the most notorious PFAS, perfluorooctanoic acid (PFOA). This is huge news. PFOA, once used in products like non-stick cookware, has long resisted degradation, earning it a spot in the infamous group of “forever chemicals.” But now, T7RdhA is poised to challenge that status.
Why Should We Care About This Protein?
You might be wondering why a single protein could make such a big difference. The answer lies in its unique ability to act as a catalyst in breaking down PFAS. Think of it like a lock and key: the protein is the lock, and the PFAS molecules are the keys. When the right key fits into the lock, it triggers a chemical reaction that starts breaking down the pollutant into less harmful components.
In the study, researchers used AlphaFold2, a revolutionary tool that predicts the 3D structure of proteins with near-experimental accuracy. This tool was pivotal in identifying how T7RdhA could bind with PFOA, setting the stage for its breakdown. The implications are profound—not just for environmental science, but for public health, too.
What is Alpha Fold 2?
AlphaFold 2 is a artificial intelligence (AI) system developed by DeepMind, a subsidiary of Alphabet (Google’s parent company), designed to predict the three-dimensional structure of proteins from their amino acid sequences with remarkable accuracy. Proteins are essential molecules that perform a wide range of functions in living organisms, and their function is heavily dependent on their 3D structure. Understanding this structure is crucial for numerous fields, including drug discovery, biotechnology, and the study of diseases.
Traditionally, determining a protein’s structure has been a labor-intensive and costly process, often requiring years of experimental work using techniques like X-ray crystallography, cryo-electron microscopy, or nuclear magnetic resonance (NMR) spectroscopy. AlphaFold 2 revolutionizes this process by using deep learning techniques to predict protein structures with a level of accuracy that rivals experimental methods.
A Game-Changer for Public Health
For decades, PFAS have been linked to a slew of health issues, including cancer, liver damage, and immune system dysfunction. They’re in our water, our soil, and even in the food we eat. Traditional methods of dealing with PFAS contamination are costly and inefficient. But if we can harness the power of proteins like T7RdhA, we could revolutionize how we approach environmental cleanup.
Imagine being able to introduce bacteria into contaminated environments, where they could naturally degrade PFAS over time. This could reduce the need for expensive and energy-intensive methods like incineration or chemical treatment. The potential cost savings and environmental benefits are enormous.
Bringing Science to Life: Why This Matters to You
You don’t have to be a scientist to appreciate the significance of this discovery. The beauty of T7RdhA lies in its potential to address a problem that affects us all—pollution. If PFAS can be broken down into less harmful substances, we could see cleaner water, healthier ecosystems, and a reduction in the health risks associated with these chemicals.
This research also underscores the power of collaboration between different fields of science. The use of AlphaFold2 to predict protein structures and functions is a perfect example of how technology and biology can come together to solve real-world problems. It reminds us that innovation often happens at the intersection of disciplines.
The Road Ahead: Challenges and Opportunities
While the discovery of T7RdhA’s capabilities is exciting, it’s important to remember that we’re still in the early stages. More research is needed to fully understand how this protein works and how we can best utilize it in real-world applications. Scaling up this technology for widespread use also presents challenges.
But the potential rewards make these challenges worth tackling. If we can overcome these hurdles, we can see a future where PFAS is no longer a looming threat but a problem that science has solved. This would be a monumental step forward in our quest for a cleaner, healthier planet.
Join the Conversation
We’re on the brink of a new era in environmental science. How do you think we can best harness the power of proteins like T7RdhA to clean up our planet? What other applications could this technology have beyond PFAS degradation?
Let’s continue this conversation in the comments below. Your insights and ideas could help shape the future of environmental cleanup.
Join the Science Adventure
Stay updated with the latest discoveries in science! Our weekly newsletter is perfect for teachers and science enthusiasts. Get the newest research, major breakthroughs, and fascinating stories delivered to your inbox for free. Enhance your teaching and learning. Subscribe today! If you liked this blog, please share it! Your referrals help This Week in Science reach new readers.



